Assessing samples for genomic testing
Overview
The purpose of assessment
The results of genomic testing will only be as good as the quality of the tissue used for genomic testing. Pathology staff are those best equipped to assess the quality of tissue, and so they have a vital role to play in genomic testing.
Assessing samples is important for testing:
It is essential to spend time assessing the suitability of all tissue samples and all blocks to select the one which is most likely to generate the highest quality genomic results. Failure to invest effort here increases the risk of false negative results (which leaves patients receiving suboptimal treatment) and of fails (which result in delays from repeat testing).
Providing metrics on tissue cellularity and percentage area necrosis can provide explanations for failed testing and may therefore guide the strategy for further tissue acquisition.
Providing an estimate of neoplastic cell percentage is very important for interpreting negative results. If the neoplastic cell percentage is low, it is possible that a negative result could represent a false negative. It is not possible to determine which nucleic acids are derived from neoplastic versus non-neoplastic cells, and so histological assessment is the only way of determining the likelihood of a false negative result.
Choosing the best block
Various factors should be considered when choosing the most appropriate block for genomic testing.
It should have a high neoplastic cell percentage
This means: what proportion of all nuclei in the section belong to neoplastic cells? Note that a sample may have a large number of tumour cells, but its neoplastic cell percentage may be low if there are also large numbers of inflammatory or stromal cells.
The idea is that as the neoplastic cell percentage falls, nucleic acids from neoplastic cells are ‘diluted’ by nucleic acids from non-neoplastic cells. If the ‘concentration’ of neoplastic nucleic acid drops below a certain point, any genomic variants present may be missed.
The ideal sample will contain 100% neoplastic cells with no non-neoplastic cells. In reality, this virtually never happens. There will almost invariably be tumour-associated inflammatory cells and stromal cells; there may also be background non-neoplastic epithelium.
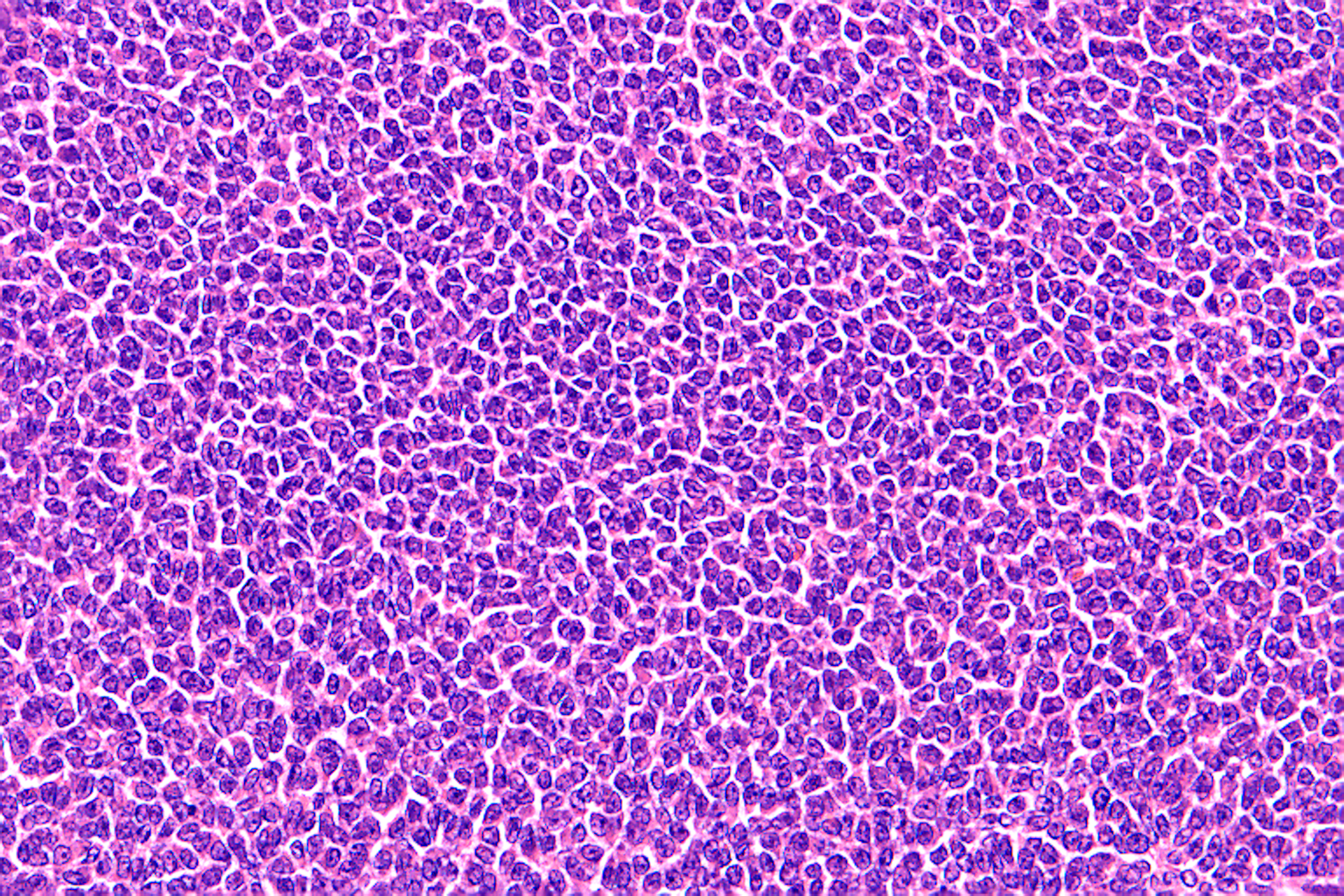
An adult granulosa cell tumour. It comprises almost entirely neoplastic cells, with essentially no admixed inflammatory or stromal cells. Neoplastic cell percentage here is 90-100%. Any variant present is highly likely be detected.
A metastatic ovarian mucinous carcinoma. There are a few malignant glands. However, there are huge numbers of lymphoid cells and substantial numbers of stromal cells. Each lymphoid cell is much small than a malignant cell, and so in even a small lymphoid aggregate there will be huge numbers of non-neoplastic cells. In this field, neoplastic cell percentage will be around 10%. Most genomic technologies require a minimum neoplastic cell content of around 20%. Therefore, if the neoplastic cells harbour a genomic variant, it may be missed, giving rise to a false negative result.
Blocks containing lymph node tissue or inflammation often have low neoplastic cell content and so are not optimal for genomic testing. Nodal metastases are therefore often not good sources of nucleic acid for genomic testing,
Blocks containing large amounts of fat may well be good for genomic testing. Adipocytes are so large that even in a sample containing huge amounts of fat, only a relatively small number of cells will be present. Therefore even very large amounts of fat are unlikely to significantly affect the neoplastic cell percentage. Equally, normal lung parenchyma often does not have a dramatic impact.
Red blood cells of course do not contribute to neoplastic cell percentage, since they do not contain nuclei.
It should contain a high absolute number of nucleated cells
Samples which contain only small numbers of nucleated cells (neoplastic or non-neoplastic) are likely to fail for genomic testing. If there are few neoplastic cells, there will be little nucleic acid in the sample. If there is insufficient nucleic acid testing will outright fail.
It is worth mentioning that modern sequencing techniques can work with quite small amounts of nucleic acid. However, different techniques have widely varying input requirements. Furthermore, samples whose nucleic acids are degraded (e.g. extended formalin fixation, decalcification) will generally require more nucleated cells to be able to obtain a successful testing result. Therefore, the ‘better’ tissue is handled from a genomic testing perspective, the less is needed.
It should contain as little necrosis as possible
There is evidence suggesting that large amounts of necrosis may be associated with an increased risk of failure. Certainly, if a sample comprises largely necrosis with only a fairly small number of viable nucleated cells, the odds of failed testing increase.
It should not be decalcified
Decalcification has a profoundly detrimental impact on nucleic acid quality. The degree of harm varies with decalcification solution used and with duration of decalcification. Nonetheless, there should be a preference for selecting for genomic testing tissue which has not been subjected to decalcification.
It should be well preserved
It is certainly true that formalin fixation degrades nucleic acids and that the greater the duration of fixation, the greater the likelihood of testing failing. However, ischaemia also results in nucleic acid degradation. Therefore, samples which show evidence of delayed or inadequate fixation may yield poorer results from genomic testing. It is wise to avoid using samples which show autolysis or poor preservation.
It should be as recent as possible
It is easy to see how nucleic acids degrade in cut sections. However, it is also known that nucleic acids degrade in paraffin blocks. After a few years there is a noticeable increase in failure rates from genomic testing, particularly for RNA sequencing. It is preferable to undertake testing on the most recent samples available.
Macrodissection
It is not uncommon for part of a section to be more suitable for genomic testing than other parts. For example, a single section may contain an area of tumour with very high neoplastic cell percentage, but at one edge may contain an uninvolved lymph node.
In these cases, it may be possible to take slide-mounted sections and physically remove the suboptimal areas of tissue from the slide, leaving only the better areas for genomic testing. This process - ‘macrodissection’ - is a means of enriching the sample for neoplastic cells. The same process may also be used to remove areas of necrosis from a section.
The pathologist’s role is to mark on an H&E-stained section the area(s) which are optimal for genomic testing. Staff in the laboratory will then use this marked H&E as a guide for macrodissection. This is particularly important for cases where the neoplastic cell percentage is lower than the minimum required for the technique; macrodissection may raise the neoplastic cell percentage above this minimum requirement and therefore avoid the risk of false negative results.
There are a few considerations to bear in mind:
It is always essential to remember that somebody will be using your marked H&E as a guide to scrape tissue from a section of unstained tissue. By its very nature, this process is not entirely accurate. It is important not to mark areas which are too small or too complex; doing so runs the risk of the dissector scraping off the area of tissue which is of high quality.
In general, GLHs prefer to receive scrolls/curls. Remember that this precludes the possibility of macrodissection. If macrodissection is required, slide-mounted sections must be prepared.
When marking an H&E there is clearly a trade-off between neoplastic cell percentage and overall cellularity. Marking a very small area may improve the neoplastic cell percentage (reducing the risk of a false negative result) but will usually markedly reduce the overall cellularity (increasing the risk of failure). This is a difficult balance, but in general it is worth emphasising that modern techniques often require rather little in the way of nucleated cells (although if the sample has been excessively fixed or decalcified, higher cellularity will be needed).
Some samples are rarely suitable for macrodissection. For example, it is rare to be able to usefully macrodissect a cell block since it will usually contain a fairly homogeneous mixture of neoplastic and non-neoplastic tissue.
Macrodissection cannot be undertaken on frozen tissue collected for WGS.
For non-WGS genomic testing, some combination of the following three metrics will be required:
Neoplastic cell percentage
Total cellularity
Percentage area necrosis
It is important to communicate clearly on the GLH request form what has been assessed. For example, an entire section may have a neoplastic cell percentage of 10% and necrosis occupying 20%, but a marked area may have a neoplastic cell percentage of 30% and necrosis occupying 5%. It is wise to communicate the assessment of the whole section and of the marked areas to the GLH clearly; in a case like this, whether or not macrodissection is performed will have a bearing on the interpretation of the final result.
For WGS, neoplastic cell percentage and percentage area necrosis are required.
Training and EQA schemes are available for tumour assessment.
See Tumour Assessment in the Genomic Era course from Health Education England →
What needs to be assessed?
Neoplastic cell percentage
The concept of neoplastic cell percentage is discussed in more detail above.
Neoplastic cell percentage is strictly an estimate; there is absolutely no need to count cells.
In some cases, it may be easier to visualise ratios than percentages. For example, it may be easier to judge whether there are four non-neoplastic cells for every neoplastic cells, than to judge whether 20% of the nucleated cells are neoplastic.
Total cellularity
This is the total number of nucleated cells (neoplastic or non-neoplastic) in a sample.
The following definitions are used by the Genomics Education Programme:
Less than 100 cells
100-4,000 cells
4,000-10,000 cells
10,000-50,000 cells
More than 50,000 cells
Very low
Low
Intermediate
High
Very high
It is worth stressing that overall cellularity is also strictly an estimate. A helpful rule of thumb is that a 20× field which is packed full of neoplastic cells will usually contain around 2,500 cells.
Percentage area necrosis
This is the percentage area of the tissue section which is occupied by necrosis.